Mahdieh Khanmohammadi
Beyond Core and Penumbra: Bi-Temporal Image-Driven Stroke Evolution Analysis
Feb 07, 2026Abstract:Computed tomography perfusion (CTP) at admission is routinely used to estimate the ischemic core and penumbra, while follow-up diffusion-weighted MRI (DWI) provides the definitive infarct outcome. However, single time-point segmentations fail to capture the biological heterogeneity and temporal evolution of stroke. We propose a bi-temporal analysis framework that characterizes ischemic tissue using statistical descriptors, radiomic texture features, and deep feature embeddings from two architectures (mJ-Net and nnU-Net). Bi-temporal refers to admission (T1) and post-treatment follow-up (T2). All features are extracted at T1 from CTP, with follow-up DWI aligned to ensure spatial correspondence. Manually delineated masks at T1 and T2 are intersected to construct six regions of interest (ROIs) encoding both initial tissue state and final outcome. Features were aggregated per region and analyzed in feature space. Evaluation on 18 patients with successful reperfusion demonstrated meaningful clustering of region-level representations. Regions classified as penumbra or healthy at T1 that ultimately recovered exhibited feature similarity to preserved brain tissue, whereas infarct-bound regions formed distinct groupings. Both baseline GLCM and deep embeddings showed a similar trend: penumbra regions exhibit features that are significantly different depending on final state, whereas this difference is not significant for core regions. Deep feature spaces, particularly mJ-Net, showed strong separation between salvageable and non-salvageable tissue, with a penumbra separation index that differed significantly from zero (Wilcoxon signed-rank test). These findings suggest that encoder-derived feature manifolds reflect underlying tissue phenotypes and state transitions, providing insight into imaging-based quantification of stroke evolution.
Self-Supervised Few-Shot Learning for Ischemic Stroke Lesion Segmentation
Mar 16, 2023Abstract:Precise ischemic lesion segmentation plays an essential role in improving diagnosis and treatment planning for ischemic stroke, one of the prevalent diseases with the highest mortality rate. While numerous deep neural network approaches have recently been proposed to tackle this problem, these methods require large amounts of annotated regions during training, which can be impractical in the medical domain where annotated data is scarce. As a remedy, we present a prototypical few-shot segmentation approach for ischemic lesion segmentation using only one annotated sample during training. The proposed approach leverages a novel self-supervised training mechanism that is tailored to the task of ischemic stroke lesion segmentation by exploiting color-coded parametric maps generated from Computed Tomography Perfusion scans. We illustrate the benefits of our proposed training mechanism, leading to considerable improvements in performance in the few-shot setting. Given a single annotated patient, an average Dice score of 0.58 is achieved for the segmentation of ischemic lesions.
Exploiting 4D CT Perfusion for segmenting infarcted areas in patients with suspected acute ischemic stroke
Mar 15, 2023Abstract:Precise and fast prediction methods for ischemic areas (core and penumbra) in acute ischemic stroke (AIS) patients are of significant clinical interest: they play an essential role in improving diagnosis and treatment planning. Computed Tomography (CT) scan is one of the primary modalities for early assessment in patients with suspected AIS. CT Perfusion (CTP) is often used as a primary assessment to determine stroke location, severity, and volume of ischemic lesions. Current automatic segmentation methods for CTP mostly use already processed 3D color maps conventionally used for visual assessment by radiologists as input. Alternatively, the raw CTP data is used on a slice-by-slice basis as 2D+time input, where the spatial information over the volume is ignored. In this paper, we investigate different methods to utilize the entire 4D CTP as input to fully exploit the spatio-temporal information. This leads us to propose a novel 4D convolution layer. Our comprehensive experiments on a local dataset comprised of 152 patients divided into three groups show that our proposed models generate more precise results than other methods explored. A Dice Coefficient of 0.70 and 0.45 is achieved for penumbra and core areas, respectively. The code is available on https://github.com/Biomedical-Data-Analysis-Laboratory/4D-mJ-Net.git.
Multi-input segmentation of damaged brain in acute ischemic stroke patients using slow fusion with skip connection
Mar 18, 2022
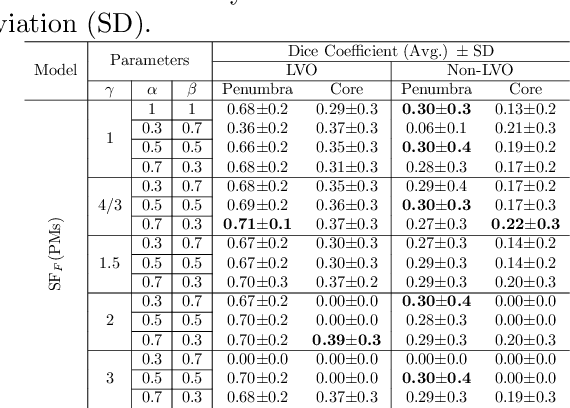
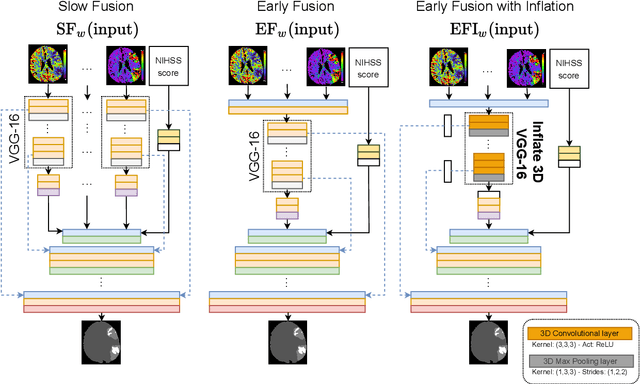
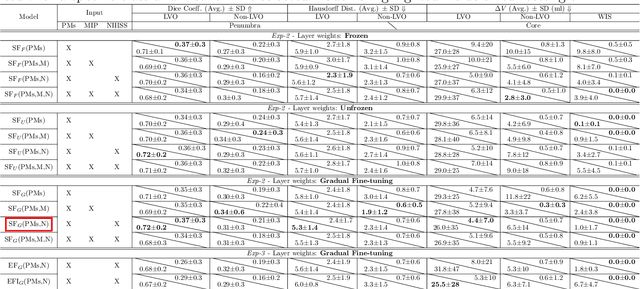
Abstract:Time is a fundamental factor during stroke treatments. A fast, automatic approach that segments the ischemic regions helps treatment decisions. In clinical use today, a set of color-coded parametric maps generated from computed tomography perfusion (CTP) images are investigated manually to decide a treatment plan. We propose an automatic method based on a neural network using a set of parametric maps to segment the two ischemic regions (core and penumbra) in patients affected by acute ischemic stroke. Our model is based on a convolution-deconvolution bottleneck structure with multi-input and slow fusion. A loss function based on the focal Tversky index addresses the data imbalance issue. The proposed architecture demonstrates effective performance and results comparable to the ground truth annotated by neuroradiologists. A Dice coefficient of 0.81 for penumbra and 0.52 for core over the large vessel occlusion test set is achieved. The full implementation is available at: https://git.io/JtFGb.
CNN Based Segmentation of Infarcted Regions in Acute Cerebral Stroke Patients From Computed Tomography Perfusion Imaging
Apr 21, 2021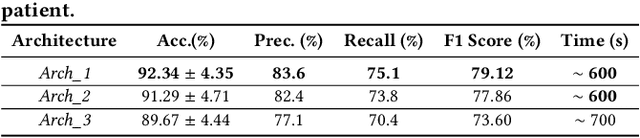
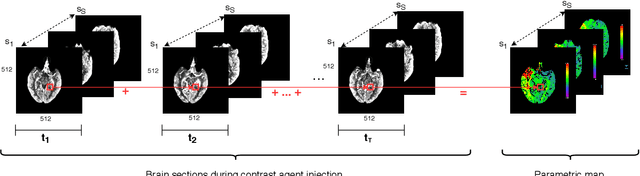
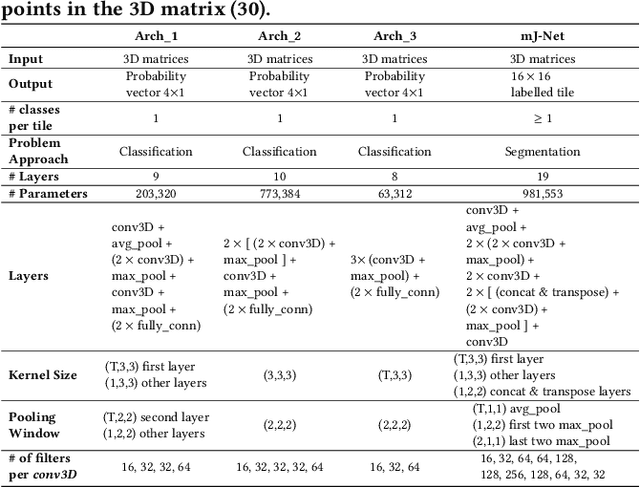
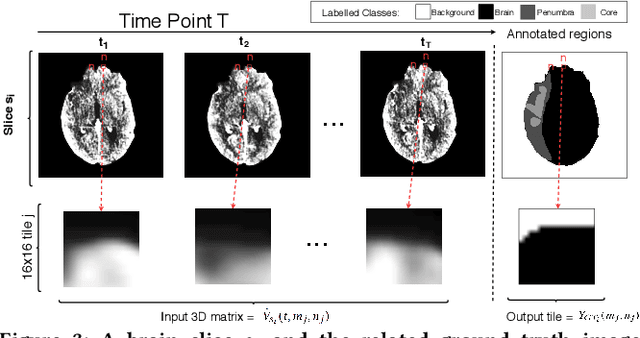
Abstract:More than 13 million people suffer from ischemic cerebral stroke worldwide each year. Thrombolytic treatment can reduce brain damage but has a narrow treatment window. Computed Tomography Perfusion imaging is a commonly used primary assessment tool for stroke patients, and typically the radiologists will evaluate resulting parametric maps to estimate the affected areas, dead tissue (core), and the surrounding tissue at risk (penumbra), to decide further treatments. Different work has been reported, suggesting thresholds, and semi-automated methods, and in later years deep neural networks, for segmenting infarction areas based on the parametric maps. However, there is no consensus in terms of which thresholds to use, or how to combine the information from the parametric maps, and the presented methods all have limitations in terms of both accuracy and reproducibility. We propose a fully automated convolutional neural network based segmentation method that uses the full four-dimensional computed tomography perfusion dataset as input, rather than the pre-filtered parametric maps. The suggested network is tested on an available dataset as a proof-of-concept, with very encouraging results. Cross-validated results show averaged Dice score of 0.78 and 0.53, and an area under the receiver operating characteristic curve of 0.97 and 0.94 for penumbra and core respectively
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge